How we Built the Ingestion Framework

Extracting Metadata from Every Corner: The evolution of the Ingestion Framework at OpenMetadata
How we built the Ingestion Framework
The first step for any user of OpenMetadata is to connect to their sources. We are talking about an ever-growing list of currently 60 services that we need to extract metadata from, translate it to the OpenMetadata standard, and send it to the server.
Without metadata, there are no discovery, collaboration, or quality tests. The ingestion process is a requirement that unlocks the rest of the features, and we are constantly pushing for improvements.
In the last year, two major transformations have helped us bring the Ingestion Framework to the next level. This post will describe the tools, decisions, and designs that made the Ingestion Framework simpler, more robust, and easier to evolve.
High-Level Design

High-level design: Source, Process, and Sink
At OpenMetadata, every action is an API call, and the Ingestion Framework is no different.
A Python package defines the process that loads metadata into the OpenMetadata server. Of course, many pieces and utilities are involved, such as the OpenMetadata client — a typed wrapper of the server APIs. Still, if we stick to the core ideas, we need to answer a few questions:
How do we interact with all the different sources?
How do we generalize and centralize the process and communications with the server?
How do we allow this process to run anywhere?
The first question is essential but does not impact the design nearly as much as the other two: We need a generic module to help us grow the list of supported sources while not tying us to specific orchestration tools.
Separation of Concerns
We achieved a method that’s generic enough by defining a Workflow class as a step-wise process that simplifies and moves data from a Source into a Sink. This allows us to put the scope on pieces that are independent of each other.
Thanks to the OpenMetadata standard, each Source only needs to focus on how to extract and translate metadata but does not really care about what happens next. In the same line of thought, the Sink class will only handle how the metadata is sent to the OpenMetadata server.
The Workflow class can then be created dynamically based on a JSON structure specifying the Source, Sink, and how to connect to the OpenMetadata Server.
{
"source": { … },
"sink": { … },
"workflowConfig": { … }
}
Therefore, any orchestrator with a Python environment can load such a JSON and run the workflow definitions from the openmetadata-ingestion package.
Simple is Better than Complex
After we defined an ingestion structure that worked, the main goal was to put it out in the open. We needed our community to have a fast and easy way to ingest metadata so they could start exploring the rest of the platform’s benefits. Thus, the ingestion team created a CLI that ran the workflow from a JSON file like the above.
However, it was not sustainable. For a few releases, the number of issues and questions related to the ingestion was never-ending. We spilled internal logic for the users to understand and relied on their laptops or host environments to run the process. We were paying the price of forcing a complex step before users could use the rest of the tool.
Democratizing (meta)data means building products that aren’t only useful for developers: our next goal was making the ingestion process available from the UI. This would give us control over the end-to-end flow and let our users focus on what they want, not on what they need to get there.
Containers, Connections, and Dynamic DAGs
The idea was simple: the server should be the only point of contact for every interaction, including the metadata workflows. Any user should be abstracted entirely over the underlying tooling and have complete control in the UI to schedule new ingestions, check the logs, or rerun them manually.
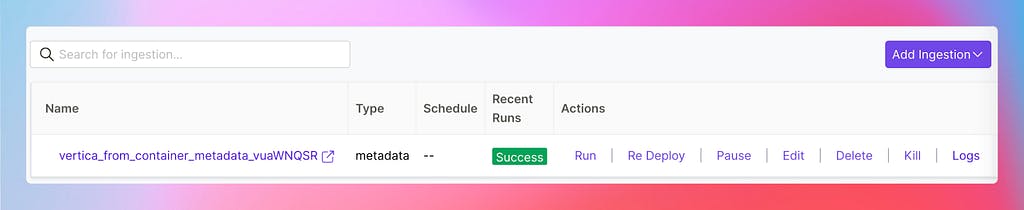
Managing Ingestion Pipelines in the OpenMetadata UI
The tool of choice for the OSS version has been Airflow, which is widely known and has already been adopted in several organizations. However, the exciting challenge here was that Airflow did not provide the main functionality we needed: deploying DAGs dynamically from an API call. So we built our own custom plugin, which is now the communication channel between Airflow and the OpenMetadata server.
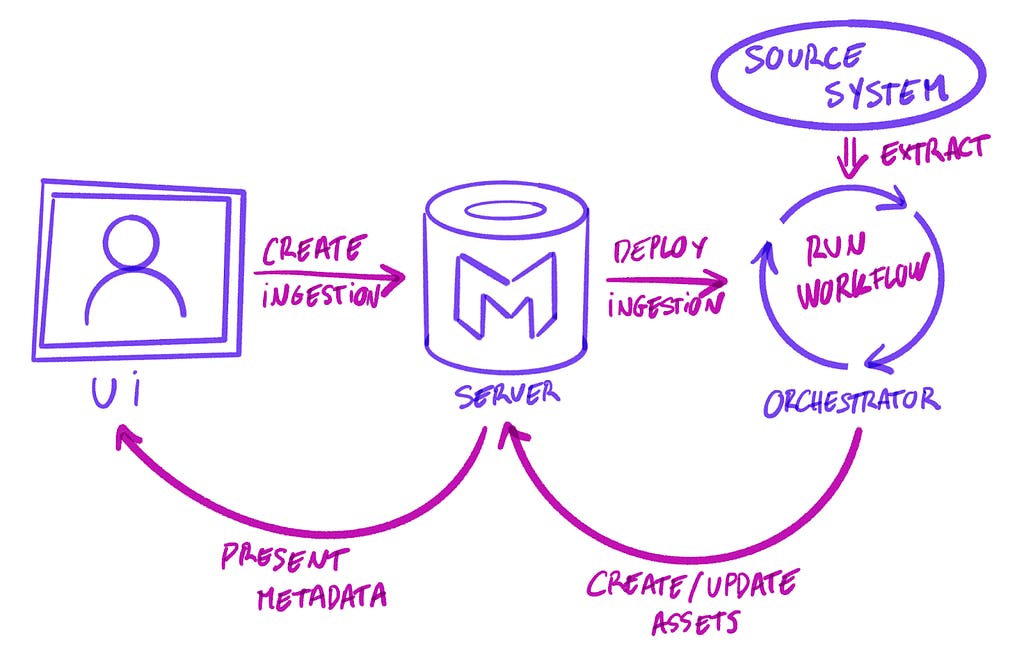
UI, Server, and Workflow interactions
Thanks to the Airflow plugins, we could define a set of APIs on top of the Ingestion Framework: from deploying a DAG to testing the connection to a service. This meant we now needed to define the requirements to connect to each service globally. The Ingestion Framework was not the only one that needed to be aware of the parameters to reach a database or connect to an external API. These connections were now filled in the UI and sent over to Airflow to be run.
As we were already using JSON Schema for the OpenMetadata standard, we migrated all the connection definitions into the specification, making them available to every part of the code. This broader use of service connections also unlocked another element to be tackled: Security. This is a topic extensive enough to require an entire post, so for now, we’re going to link to the docs for curious minds.
Service Topologies
The last piece of the puzzle was not related to the UX or architecture but to the nature of the problem at hand: How could we safely maintain and evolve a list of sources that will only grow? Moreover, as an open-source project, how can we make contributors’ lives easier?
We needed an approach checking the following boxes:
It should be flexible enough to allow us to perform generic improvements and iterations to the whole framework without many reworks.
We should standardize the requirements for each service type. We don’t have the same metadata when ingesting a database source vs. an ML Model.
Developers should have precise recipes to follow to add new sources.
Abstract classes help the interpreter notice if a specific method is missing in an implementation. Still, the extraction process has a particular order (hierarchy) that must also be followed. For example, we can only ingest a table once its schema has been processed, and the lineage of a dashboard requires the dashboard itself to be present in OpenMetadata first.
Each service must follow a series of extraction steps to be executed in a specific order. This idea was the foundation of Service Topologies: a sequence of Nodes and Stages that define in which order the metadata is extracted from the Source and which transformations are applied to produce the OpenMetadata assets.

Pipeline Service Topology
If we look at the Pipeline Topology, we will first get and create the Pipeline Service. Afterward, we will obtain the list of pipelines to be ingested and iterate over them to create the pipeline assets in OpenMetadata and send their tags, status, or lineage information.

Database Service Topology
The same thing happens when talking about Database Services. The nodes guide us through the asset hierarchy: Service -> Database -> Schema -> Table. Order matters, as we can only create a table once we know the schema it belongs to. These ingredients are then shared in the execution context: the Topology also helps us define the required information for processing each Stage.

High-level Topology description
Python dynamism played a crucial role in this solution, as we can pick up on-the-fly the methods that need to be executed at each moment. Updating the service extraction becomes as easy as adding a new abstract method to be implemented and its name in the Topology as a string at the exact position that the execution needs to happen.
Summary
In this post, we’ve had a glimpse of the evolution of the Ingestion Framework during this last year. Our major focus has been improving the UX with the UI integration of the ingestion while also focusing on the internal design to make the code easier to maintain and evolve.
Code is never finished, only abandoned, and we will keep working to make the Ingestion Framework better release after release. These improvements have been essential, but there is still room for improvements to make the ingestion faster and more robust.
If you find this interesting, want to chip in, or just say hi, do not hesitate to reach out! Please take a look at our GitHub repo, give it a star, and drop us a line in Slack 🚀

How we Built the Ingestion Framework was originally published in OpenMetadata on Medium, where people are continuing the conversation by highlighting and responding to this story.


