How to Integrate OpenMetadata Test Suites with Your Data Pipelines

With OpenMetadata v0.12.0 onwards, users can create and execute test suites directly from the user interface for Data Quality. While this approach covers most use cases, certain data teams will want to integrate their test suite executions with their data processing pipelines. Instead of validating data after it has been modeled, integrating your test suite execution with your data processing pipeline allows you to stop any further tasks to run if quality criteria are not met.
In this article, we will go over OpenMetadata test suite concepts, how to create a test suite in OpenMetadata, and how to integrate your test suite execution with your data processing pipelines. We’ll use Airflow as our workflow scheduler for our pipelines, but the concepts apply to any type of workflow.
What are OpenMetadata Test Suites
At their core, Test suites are execution containers. They hold test cases, which themselves represent the specification of test definitions. OpenMetadata provides a growing number of table-level tests [details] and column-level tests [details], as well as custom SQL tests allowing you to write SQL tests [details].
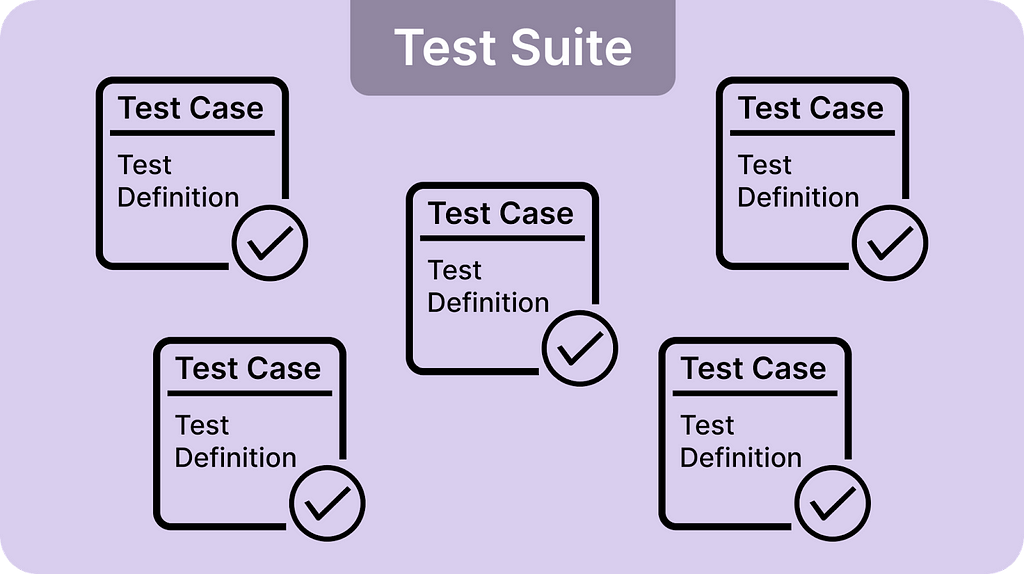
Image 1-0 — Test Suite, Test Case, and Test Definition relationship
Test Suite represents a grouping of test cases for a table. In OpenMetadata 1.1.0 and up, test suites are divided into executable (test suites used to run test cases) and logical (test suites used to group tests together and visualize the assertion results). Tests from different tables can be grouped within the same logical test suite. This allows users to visualize tests from different tables in one place. We can imagine multiple source tables used to model a downstream table. All the tests from the source tables can be grouped together to ensure the accuracy of the newly modeled table.
How to Create a Test Suite?
Make sure you have a running instance of OpenMetadata v1.1.0 or later. If you don’t, check out our Try OpenMetadata in Docker. Once OpenMetadata is running, ingest metadata for a Database service. Then, navigate to your table data asset and click “Profiler & Data Quality”. From there, you will be able to create a test. Go ahead and click on “Add Test” — you will need to select your test type “Table” or “Column”. Next, you will have to select the test definition you want to run. Select the definition you want and configure your test according to your needs.
The final step will be to add an ingestion workflow for our test execution. To integrate our test suite execution with our data processing pipeline, we won’t add a schedule. On the “Schedule for Ingestion” page, simply select “none” as a scheduled time.

Image 1.1 — Test Suite Creation
Integrating OpenMetadata in your Data Processing Pipeline
For this example, we’ll create a simple pipeline with 3 tasks (data extraction, OpenMetadata test suite, modelization). We’ll use Los Angeles Sewer data for our example. The data can be fetched at the following API endpoint 👉 https://data.lacity.org/resource/qcn5-9nfy.json. It also assumes you have:
A running OpenMetadata instance, and
Followed the instructions in “How to Create a Test Suite” to create a test suite.
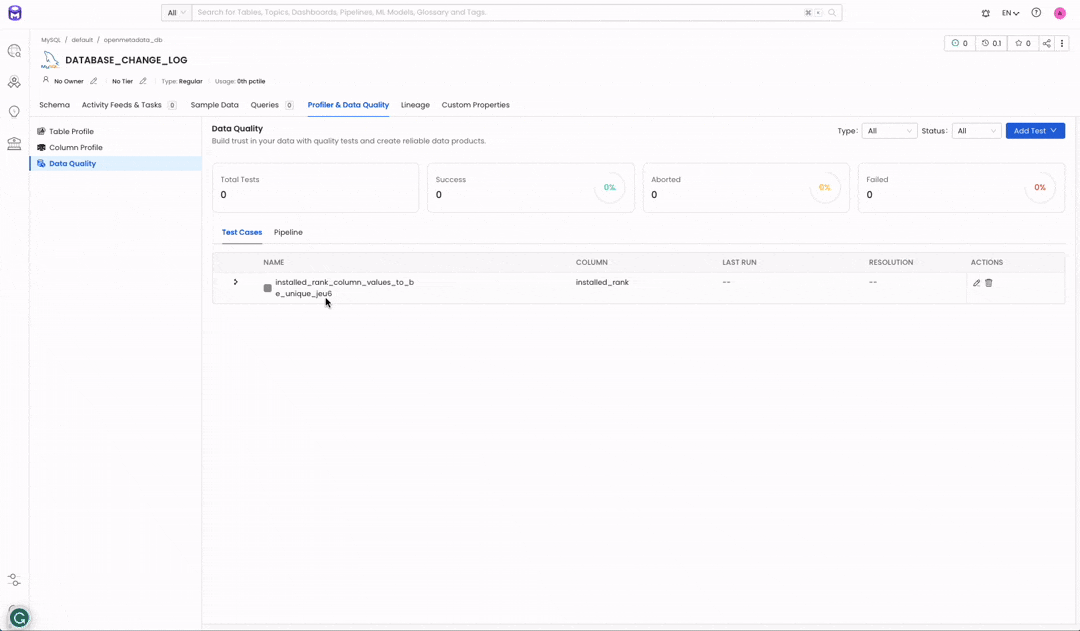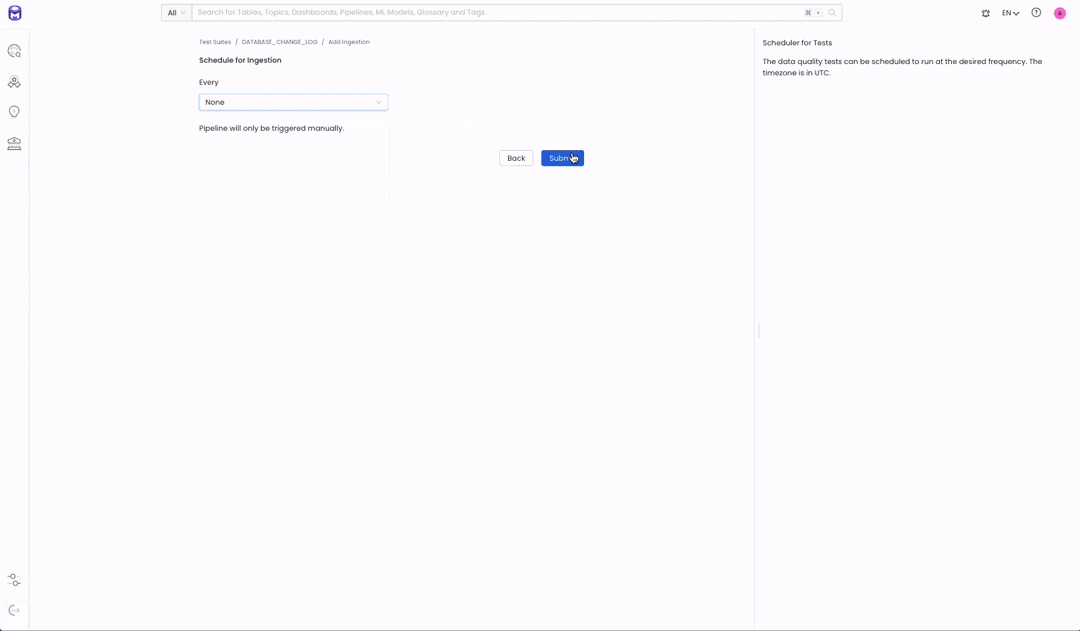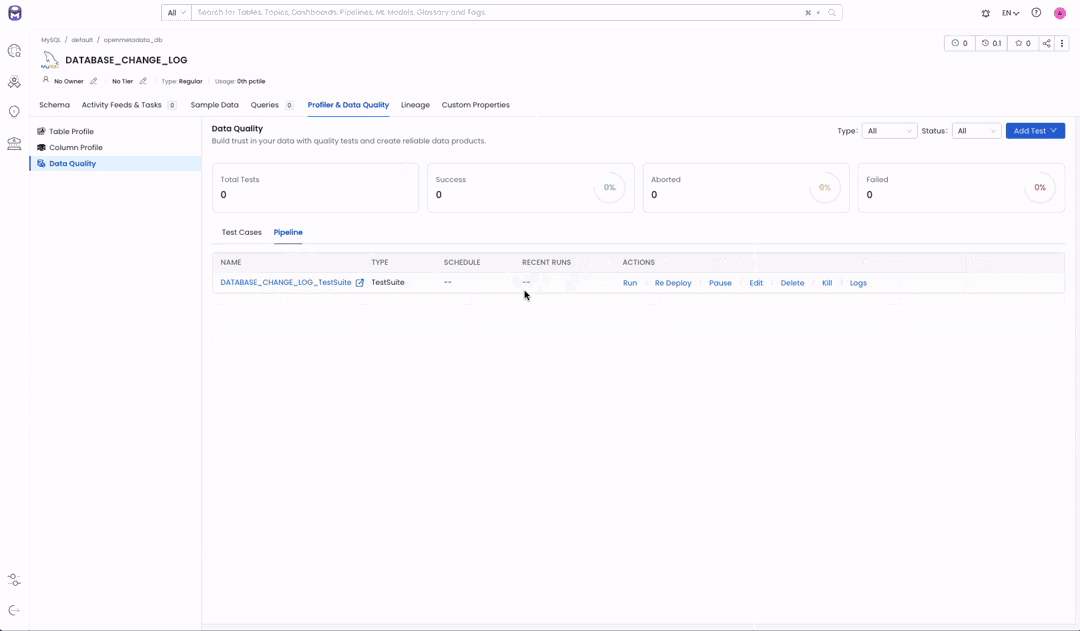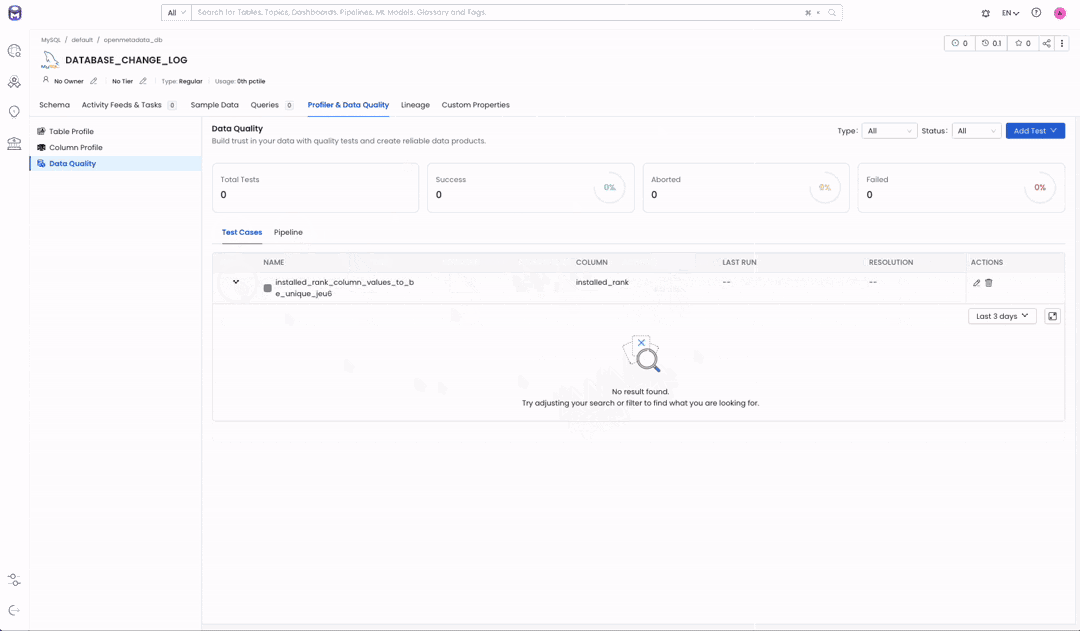
Image 1.2 — Test Suite before executing our pipeline
Extract Source Data
Example 1–1 shows our Airflow task used to extract Los Angeles sewer data using the API.
example 1–1 — extract and insert task
@task(task_id="extract_and_insert_data") ①
def extract_and_insert(**kwargs):
"""Extract function"""
resp = requests.get(ENDPOINT)
df = pd.DataFrame(resp.json())
hook = RedshiftSQLHook() ②
engine = hook.get_sqlalchemy_engine() ③
engine.dialect.description_encoding = None
df.iloc[:100,1:].to_sql(
"los_angeles_sewer_system",
con=engine,
if_exists="replace",
index=False,
) ④
① @task is an Airflow decorator used to define tasks in the pipeline
② we get a DbpApiHook which allows us to access specific SQLAlchemy methods
③ we get the SQLAlchemy engine (our database connection) that we will use to write our data
④ write 100 rows skipping the first column
Now that we have written our source extraction function, the next step is to execute our OpenMetadata tests against our data source. Before moving on to the next step, ensure that you have:
Ingested metadata for the source table, and
Created a test suite with some tests for your data source table (we named our test suite LASewerTests).
Execute Tests Against the Source Data
We chose to break down this step into 2 tasks, although it can be combined together. First, we will execute the test suite (i.e. run the test cases associated with our test suite) (example 1–3) and then we will check the results of our test suite’s test cases (example 1–4). We also defined a get_ingestion_pipeline() function (example 1–2) used by 1–3 and 1–4.
example 1–2 — get ingestion pipeline util function
def get_ingestion_pipeline(name: str):
"""get ingestion pipeline by name
Args:
name (str): start or full name of ingestion
"""
pipelines = OPENMETADATA.list_entities(
IngestionPipeline,
fields="*",
).entities ①
test_suite_pipeline: Optional[IngestionPipeline] = next(
(pipeline for pipeline in pipelines if pipeline.fullyQualifiedName.__root__.startswith("LASewerTests")),
None
) ②
return test_suite_pipeline
① we use OpenMetadata Python SDK to list all of our ingestion pipelines
② we filter the test suite ingestion we have created when creating our test suite or we return None if we don’t find our test suite ingestion pipeline
example 1–3 — execute test suite pipeline
@task(task_id="run_om_test")
def run_om_test(**kwargs):
"""Run Openmetadata tests"""
test_suite_pipeline: Optional[IngestionPipeline] = get_ingestion_pipeline("LASewerTests") ①
if not test_suite_pipeline:
raise RuntimeError("No pipeline entity found for {test_suite_pipeline}")
OPENMETADATA.run_pipeline(test_suite_pipeline.id.__root__) ②
timeout = time.time() + TIMEOUT ③
while True:
statuses = OPENMETADATA.get_pipeline_status_between_ts(
test_suite_pipeline.fullyQualifiedName.__root__,
get_beginning_of_day_timestamp_mill(),
get_end_of_day_timestamp_mill(),
) ④
if statuses:
status = max(statuses, key=operator.attrgetter("timestamp.__root__")) ⑤
if status.pipelineState in {PipelineState.success, PipelineState.partialSuccess}:
break
if status.pipelineState == PipelineState.failed:
raise RuntimeError("Execution failed")
if time.time() > timeout:
raise RuntimeError("Execution timed out")
① get our pipeline entity
② run our pipeline entity which will run the tests
③ we’ll be checking the status of our pipeline using a while loop so we’ll add a timeout to make sure we don’t get stuck in an infinite loop
④ retrieve the pipeline statuses for today. get_pipeline_status_between_ts takes 2 timestamps milliseconds as its second and third argument
⑤ get the latest pipeline status
Once our pipeline finishes without failure we need to check the results of our test cases. We’ll do that in the below task.
@task(task_id="check_om_test_results")
def check_om_test_results(**kwargs):
"""Check test results"""
test_cases = OPENMETADATA.list_entities(
TestCase,
fields="*",
) ①
test_cases = [
entity for entity in test_cases.entities if entity.testSuite.name == "LASewerTests"
] ②
test_suite_pipeline: Optional[IngestionPipeline] = get_ingestion_pipeline("LASewerTests")
pipelines_statuses = OPENMETADATA.get_pipeline_status_between_ts(
test_suite_pipeline.fullyQualifiedName.__root__,
get_beginning_of_day_timestamp_mill(),
get_end_of_day_timestamp_mill(),
)
if not pipelines_statuses:
raise RuntimeError("Could not find pipeline")
latest_pipeline_status: PipelineStatus = max(pipelines_statuses, key=operator.attrgetter("timestamp.__root__"))
for test_case in test_cases:
timeout = time.time() + TIMEOUT
while True:
test_case_results = OPENMETADATA.get_test_case_results(
test_case.fullyQualifiedName.__root__,
get_beginning_of_day_timestamp_mill(),
get_end_of_day_timestamp_mill()
) ③
if time.time() > timeout:
raise RuntimeError("Execution timed out")
if test_case_results:
latest_test_case_result = max(test_case_results, key=operator.attrgetter("timestamp.__root__"))
if latest_test_case_result.timestamp.__root__ < latest_pipeline_status.startDate.__root__: ④
continue
if latest_test_case_result.testCaseStatus in {TestCaseStatus.Failed, TestCaseStatus.Aborted}: ④
raise RuntimeError(f"Test case {test_case.name.__root__} returned status {latest_test_case_result.testCaseStatus.value} ")
break
① Get our test case entities
② Filter test cases that belong to our test suite. It is also possible to pass a testSuiteId parameter in ①, though we prefer this approach in this demo as we have a small number of test cases, and it prevents us from sending another API request
③ We iterate through all of our test cases and retrieve the results
④ If our result timestamp is earlier than our pipeline execution, we keep iterating. This means our results are not yet available.
⑤ If our test result has either failed or was aborted, we fail our task.
Examples 1–3 and 1–4 allow us to stop our data processing if an error is raised in one of our test cases. Unlike traditional data quality tools test results will be centralized back inside OpenMetadata, allowing the whole team to know a test has failed. On top of centralizing data test failure, any user with the right permission can add additional tests to that test suite, which will be executed as part of the data processing pipeline.
It is simple to imagine a flow where a Data Engineer creates a data processing pipeline, where an OpenMetadata test suite is run, and a business user defines test cases directly from OpenMetadata user interface. This ensures that teams with business knowledge can easily define quality rules that a data asset needs to have.
Create our Modeled Data
Our final step is to model our data. Example 1–5 takes the source data and performs some aggregation.
Example 1–5
@task(task_id="model")
def model(**kwargs):
"""model function"""
hook = RedshiftSQLHook()
engine = hook.get_sqlalchemy_engine()
engine.dialect.description_encoding = None
df = pd.read_sql(
"SELECT mapsheet, avg(shape_leng) mean_shape_leng FROM los_angeles_sewer_system GROUP BY 1;",
con=engine
)
df.to_sql("sewer_mean_len", con=engine, index=False, if_exists="replace")
Conclusion
Now that we have our whole DAG it is time to execute it. As we saw in Image 1.2, OpenMetadata had no test results for our “LA Sewer Tests” test suite. We created our test case to make it fail to show how our modelization task will not run.
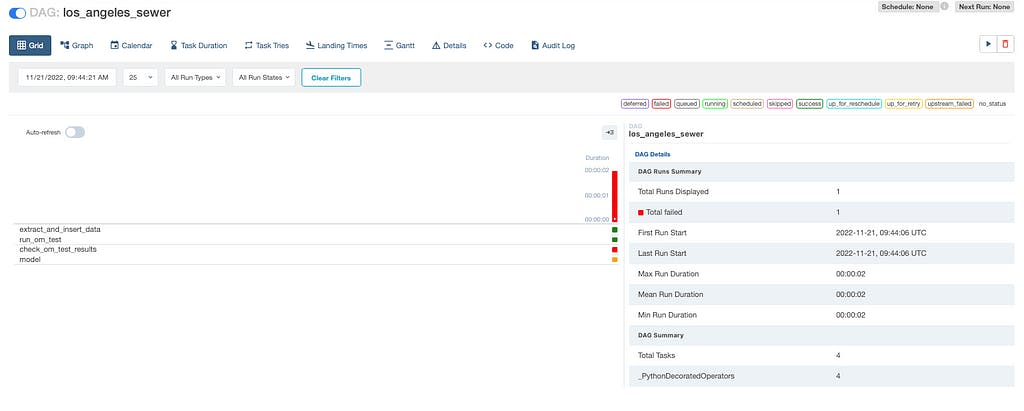
Image 1.3 — Airflow data processing pipeline execution
Image 1.3 shows the 4 tasks we previously defined. Task #4 was not executed as task #3 check_om_test_results reported test case failures and failed the Airflow task. This prevented our modelization task to run.
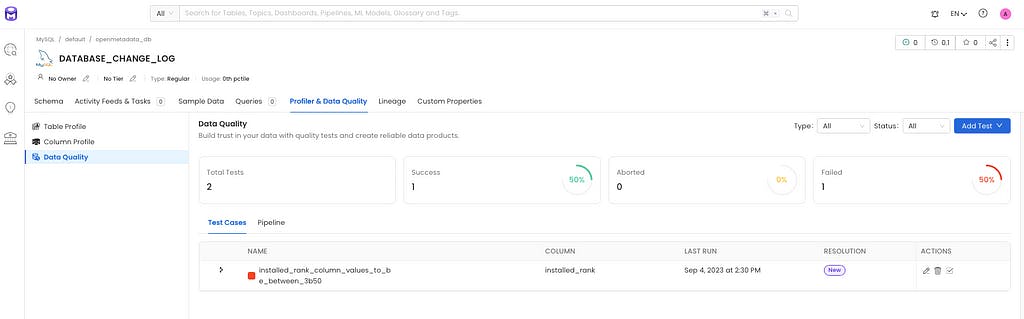
Image 1.4 — OpenMetadata UI after Airflow DAG execution
After our data processing pipeline execution, we can navigate to OpenMetdata test suite to view that our test result has been updated in our test suite with a failed test. This allows easy discovery of test failure for OpenMetadata users.
This illustrates the strength of OpenMetadata. With the API at the core of its architecture, technical teams, and business teams can easily collaborate on building accurate and reliable data products.

How to Integrate OpenMetadata Test Suites with Your Data Pipelines was originally published in OpenMetadata on Medium, where people are continuing the conversation by highlighting and responding to this story.


